|

May 2010
from
ScienceMag Website
Introduction
Recent advances in high-throughput DNA sequencing have provided
initial glimpses of the nuclear genome of Neanderthals as well as
other ancient mammals including cave bears and mammoths.
In the 7
May 2010 issue of Science, an international team of researchers
presents the draft sequence of the Neanderthal genome composed of
over 3 billion nucleotides from three individuals.
Because Neanderthals are much closer kin to us than are chimpanzees,
which diverged from the human lineage 5 to 7 million years ago,
matching Neanderthal DNA against our own has the potential to reveal
genetic changes that help define who we are.
About Neanderthals
Neanderthals (Homo neanderthalensis) are currently believed to be
our closest evolutionary relatives.
Although some researchers once
thought they were our immediate ancestors in Europe, most now agree
that Neanderthals and modern humans most likely shared a common
ancestor within the last 500,000 years, possibly in Africa.
The morphological features typical of Neanderthals first appear in
the European fossil record about 400,000 years ago, with bones of
full-fledged Neanderthals showing up at least 130,000 years ago.
They lived in Europe and western Asia,
as far east as southern Siberia and as far south as the Middle East
(see below map), before disappearing from the fossil record about
30,000 years ago.
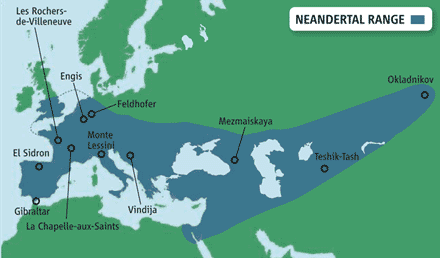
At home in Eurasia. Neanderthals ranged from Europe to southern
Siberia.
Fossil remains and anatomical
reconstructions indicate that the typical Neanderthal had a stocky
muscular body with short forearms and legs, a large head with bony
brow ridges and a brain slightly larger than ours, a jutting face
with a large nose, and perhaps reddish hair and fair skin.
Neanderthals made and used a diverse set of sophisticated tools,
controlled fire, organized their living spaces, hunted and fed on
game of various sizes, and occasionally made symbolic or ornamental
objects.
For more Neanderthal basics, see
A Neanderthal Primer by M. Balter,
Science 323, 870 (2009).
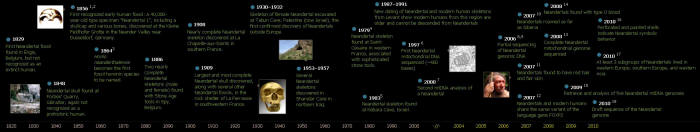
Timeline of Discoveries
The first Neanderthal fossils were discovered in 1829 in Engis,
Belgium, and in 1848 at Forbes' Quarry, Gibraltar, but were not
recognized as an early human species until after the 1856 discovery
of "Neanderthal 1" - a 40,000-year-old specimen, including a
skullcap and various bones, found at the
Kleine Feldhofer Grotte in
the
Neander Valley near Düsseldorf, Germany.
This timeline highlights key discoveries about our closest
relatives, from early fossil finds to the publication of the draft
nuclear genome sequence.
Timeline References
1. - J. C. Fuhlrott, Verh.
naturhist. Ver. preuss. Rheinl. 14, Corr. Bl., 50. (1857)
2. - H. Schaaffhausen, Verh. naturhist. Ver. preuss. Rheinl.
14, Corr. Bl., 50–52. (1857)
3. - W. King, Quarterly Review of Science 1, 88 (1864).
4. - Lévêque and Vandermeersch, Bulletin de la Société
Préhistorique Francaise 77, 35 (1980).
5. - Rak and Arensburg, Am. J. Phys. Anthropol. 73, 227
(1987).
6. - Krings et al., Cell 90, 19 (1997).
7. - Ovchinnikov et al., Nature 404, 490 (2000).
8. - Noonan et al., Science 314, 1113 (2006).
9. - Green et al., Nature 444, 330 (2006).
10. - Krause et al., Nature 449, 902 (2007).
11. - Lalueza-Fox et al., Science 318, 1453 (2007).
12. - Krause et al., Curr. Biology 17, 1908 (2007).
13. - Green et al., Cell 134, 416 (2008).
14. - Lalueza-Fox et al., BMC Evol. Biol. 8, 342 (2008).
15. - Briggs et al., Science 325, 318 (2009)
16. - Zilhao et al., Proc. Natl. Acad. Sci. U.S.A. 107, 1023
(2010).
17. - Fabre et al., PLoS ONE 4, e5151.
doi:10.1371/journal.pone.0005151 (2010).
18. - Green et al., Science 328, 710 (2010).
Methodology
The Samples
To sequence the Neanderthal genome, Green et al. began by analyzing
21 Neanderthal bones from
Vindija Cave in Croatia. The team removed
just 50 to 100 mg of bone powder from each bone and screened each
sample for the presence of Neanderthal mitochondrial DNA (mtDNA) by
PCR.
Three of the bones, which were determined to represent three
different female individuals, were ultimately chosen for sequencing
analysis.

-
The first bone dates to about 38,000 years ago and has been used for
earlier genome sequencing efforts, including determination of a
complete mitochondrial DNA sequence.
-
The second bone is undated but
was found in an older stratigraphic layer than the first bone.
-
The
third bone has been dated to about 44,500 years ago.
Ancient DNA Challenges
Extracting and analyzing ancient DNA is fraught with challenges.
Genetic material degrades into small fragments over time, with
errors often introduced into the aging sequence. Moreover, 95 to 99%
of fossil DNA can be sequences from microbes that have infiltrated
the decaying bone.
Ancient human remains pose additional challenges
in that well-preserved samples are rare and have typically been
handled by curators and researchers, thus raising the possibility
that they have been contaminated by modern human DNA. Because
Neanderthals and humans are so closely related, distinguishing
between their DNA can be nearly impossible.
Several precautions were taken in Green et al. to minimize
contamination during the Neanderthal genome project including:
-
DNA
extraction procedures performed in a "clean room" with sterile
equipment
-
the addition of unique sequence tags to ancient DNA
molecules in the clean room, making it possible to identify
contamination from other DNA sources
-
the use of enzymes that
preferentially cut microbial DNA to increase the relative
proportion of Neanderthal DNA in the sequencing libraries
Sequencing Approach
High-throughput sequencing technologies and advances in
metagenomic
analysis of complex DNA mixtures have enabled the recovery of
genomic sequences from ancient samples on a budget and timescale not
previously achievable.
To decipher the Neanderthal genome, Green et
al. used an approach known as pyrosequencing, which enables the
sequencing of hundreds or thousands of DNA molecules at the same
time, thus generating smaller pieces of sequences faster and cheaper
than classical Sanger sequencing.
One of the limitations of this
technique is that the length of DNA that can be read in any single
sequencing run is very short. For the Neanderthal genome, these
short sequences were then assembled using a combination of
sophisticated alignment algorithms as well as the completed human
and chimpanzee genome sequences as guides.
This has resulted in
~1.3-fold coverage of the entire Neanderthal genome.
Comparison of Neanderthal and present-day human genomes can reveal
information about genetic changes that have occurred before and
after the ancestral population split of modern humans and
Neanderthals (see the Comparative Genomics section for more).
However, low coverage sequencing inevitably leaves a substantial
proportion of the genome uncovered.
To recover additional
information about specific regions of interest, Burbano et al. used
a microarray-based approach to sequence ~14,000 protein-coding
regions inferred to have changed in the human lineage since the last
common ancestor shared with chimpanzees. By generating the sequence
of one Neanderthal and 50 present-day humans at these positions, the
researchers identified 88 amino acid substitutions that have become
fixed in humans since our divergence from the Neanderthals (see
table).
Further studies will be needed to investigate the possible
functional significance of these genetic changes.
Examples of genes
in which amino acid substitutions have become
fixed in humans
since our divergence from Neanderthals
|
Gene Symbol |
Gene
Name |
Molecular Function |
|
ABCC12 |
ATP-binding
cassette, subfamily C (CFTR/MRP), member
12 |
ATP-binding
cassette (ABC) transporter |
|
CASC5 |
cancer
susceptibility candidate 5 |
-- |
|
KCNH8 |
potassium
voltage-gated channel, subfamily H (eag-related),
member 8 |
Voltage-gated potassium channel |
|
LYST |
lysosomal
trafficking regulator |
Select
regulatory molecule |
|
MCHR2 |
melanin-concentrating hormone receptor 2 |
G-protein
coupled receptor |
|
OR5K4 |
olfactory
receptor, family 5, subfamily K, member
4 |
-- |
|
PNLIP |
pancreatic
lipase |
Lipase |
|
SPAG17 |
sperm
associated antigen 17 |
-- |
|
STAB1 |
stabilin 1 |
Extracellular matrix structural protein |
|
ZNHIT2 |
zinc finger,
HIT type 2 |
Zinc finger
transcription factor |
|
Comparative
Genomics
Human-Neanderthal Comparisons
Comparisons of the human genome to the genomes of Neanderthals and
apes can help identify features that set modern humans apart from
other hominin species.
In particular, the Neanderthal genome
sequence can now be used to catalog changes that have become "fixed"
(are invariant within a population or species) in modern humans
during the last few hundred thousand years and should be helpful for
identifying genes affected by positive selection since humans
diverged from Neanderthals.
To help make informative comparisons, Green et al. sequenced the
genomes of 5 present-day individuals from different parts of the
world:
They then compared the genomes of these
individuals with the genomes of the Neanderthal and the chimpanzee,
and looked both for specific regions shared by present-day humans
but lacking in Neanderthals and regions showing high frequencies of
more recently evolved (derived) sequences.
These regions signal the
presence of mutations that occurred and swept to either high
frequency or fixation after humans and Neanderthal diverged, and
that may have contributed to modern human-specific traits.
Using this comparative approach, Green et al. came up with a list of
20 candidate regions that may have been affected by positive
selection in ancestral modern humans.
Five of these regions contain
no protein-coding genes and may thus include structural or
regulatory elements. Among the remaining 15 regions, the team
identified genes involved in metabolism and cognitive and cranial
development, which suggests that aspects of these processes may have
been functionally important for the evolution of modern humans.
For more on Neanderthal-modern human comparisons see the
News story
by A. Gibbons.
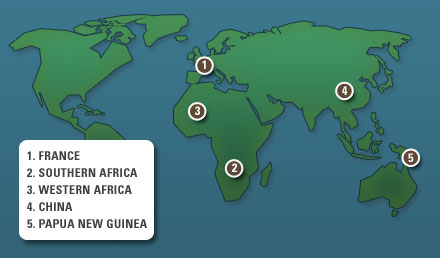
Five modern human genomes.
Locations of present-day individuals
whose genomes were sequenced
by Green et al. for comparative
analyses.
Evidence of Admixture
Substantial controversy surrounds the question of
whether
Neanderthals interbred with modern humans.
To address this question,
Green et al. tested whether Neanderthals are more closely related to
some present-day humans than to others.* Because modern humans are
believed to have originated in Africa, if Neanderthals diverged from
modern humans before present-day populations began to differentiate,
one would expect Neanderthal sequences to match sequences from
non-Africans and Africans to the same extent.
* For a description of additional methods used by Green et al. to
detect gene flow between Neanderthals and modern humans, see the
News story by A. Gibbons.
Unexpectedly, the
researchers found that Neanderthals share more genetic variants with
present-day non-Africans than with Africans. These results can be
explained if gene flow occurred from Neanderthals into the ancestors
of non-Africans.
The observation that the Neanderthal genome appears as closely
related to the genome of a Chinese and a Papua New Guinean
individual as to the genome of a French individual is particularly
surprising as there is, to date, no fossil evidence that
Neanderthals existed in East Asia or Papua New Guinea. Green et al.
thus suggest that gene flow between Neanderthals and modern humans
occurred prior to the divergence of European and Asian populations.
Based on comparative genomic data, as well as a mathematical model
of gene flow, the authors further estimate that between 1 and 4% of
the genomes of people in Eurasia may be derived from Neanderthals.
Implications for Modern Human Origins
There are two major competing hypotheses about the origins of modern
humans.
-
The "Out of Africa" hypothesis posits that modern humans
evolved from a small population in Africa and replaced all other hominin populations, including Neanderthals, as they migrated into
Europe and Asia. The simplest form of this model assumes no
interbreeding between modern and ancestral human populations.
-
In
contrast, the "Multiregional" hypothesis holds that modern humans
evolved in several regions of the world simultaneously. According to
this view, archaic humans were not replaced by anatomically modern
humans, but rather, gene flow between Africa, Europe, and Asia, led
to the evolution of modern humans from local populations.
The finding that Neanderthals are on average closer to individuals
in Eurasia than to individuals in Africa thus presents a challenge
to the strictest version of the "Out of Africa" model. However
variations of this model are plausible.
Green et al. suggest that
mixing of early modern humans ancestral to present-day non-Africans
with Neanderthals is likely to have occurred in the Middle East
prior to their expansion into Eurasia.
The authors contend that this
scenario is compatible with the archaeological record, which shows
that modern humans appeared in the Middle East before 100,000 years
ago while the Neanderthals existed in the same region after this
time, perhaps until 50,000 years ago.
Although the Green et al. analyses are suggestive of admixture, the
role of Neanderthals in the genetic ancestry of humans outside of
Africa was likely relatively minor given that only a few percent of
the genomes of present-day people outside of Africa appear to be
derived from Neanderthals.
More fossil and genetic data will help
researchers further resolve the relationships between our early
ancestors and how they shaped modern human evolution.
Neanderthals
...Didn't
Mate With Modern Humans, Study Says
by Ker Than
August 12, 2008
from
NationalGeographic Website
Neanderthals and anatomically modern
humans likely did not interbreed, according to a new DNA study.
The research further suggests that small population numbers helped
do in our closest relatives. Researchers sequenced the complete
mitochondrial genome - genetic information passed down from mothers
- of a 38,000-year-old Neanderthal thighbone found in a cave in
Croatia. (Get the
basics on genetics.)
The new sequence contains 16,565 DNA bases, or "letters,"
representing 13 genes, making it the longest stretch of Neanderthal
DNA ever examined.

Neanderthals are
depicted as cannibals in Krapina, northern Croatia,
about 32,000 years
ago in this undated illustration.
A DNA study published in August 2008 says modern humans likely did
not interbreed with Neanderthals
and that a limited
gene pool might have contributed to the demise of modern humans'
closest relatives.
Image from Mansell/Time Life Pictures/Getty Images
Mitochondrial DNA (mtDNA) is easier to
isolate from ancient bones than conventional or "nuclear" DNA -
which is contained in cell nuclei - because there are many
mitochondria per cell.
"Also, the mtDNA genome is much
smaller than the nuclear genome," said study author Richard
Green of the Max Planck Institute for Evolutionary Biology
in Germany.
"That's what let us finish this
genome well before we finish the nuclear genome," he said.
The new findings are detailed in the
August 8 issue of the journal Cell.
A Small
Population
The new analysis suggests the last common ancestor of modern
humans (Homo sapiens) and Neanderthals lived between 800,000 and
520,000 years ago. This is consistent with
previous work on shorter stretches
of Neanderthal DNA.
Contrasted with modern humans, Neanderthals exhibited a greater
number of letter substitutions due to mutations in their
mitochondrial DNA, although they seem to have undergone fewer
evolutionary changes overall.
The fact that so many mutations - some of which may have been
harmful - persisted in the Neanderthal genome could indicate the
species suffered from a limited gene pool. This might be because the
Neanderthal population was smaller than that of Homo sapiens living
in Europe at the time.
A small population size can,
"diminish the power of natural
selection to remove slightly deleterious evolutionary changes,"
Green said.
The researchers estimate the Neanderthal
population living in Europe 38,000 years ago never reached more than
10,000 at any one time.
"This could have been a factor in
their demise", Green said.
Homo neanderthalis first appeared
in Europe about 300,000 years ago but mysteriously vanished about
35,000 years ago, shortly after the arrival of modern humans -
Homo sapiens - in Europe.
"If there were only a few, small
bands of Neanderthals, barely hanging on, then any change to
their way of life could have been enough to drive them to
extinction," Green said.
"One obvious change would have been the introduction of another
large hominid - modern humans."
Stepping
Forward
Stephen Schuster, a molecular biologist at Pennsylvania State
University, said the new study should silence a lot of theories
about
interbreeding between Neanderthals and modern
humans.
The study shows that,
"at least for the maternal lineage,
there are no traceable genetic markers that suggest admixture of
Neanderthals and modern humans," he said.
Schuster added that the researchers were
exceptionally careful to isolate the Neanderthal DNA.
"Many more precautions were taken to
ensure that no contamination with human DNA has flawed the
analysis," he said, noting that researchers sequenced each
letter about 35 times to be sure of their work.
"This was the weak point of previous reports," said Schuster,
who was not involved with the study.
Thomas Gilbert, an ancient DNA
expert at the University of Copenhagen in Denmark who also was not
involved in the study, called the research a "step forward" and a
taste of what might come when the Neanderthal nuclear DNA is
finished.
The team's argument that the Neanderthal population was small 38,000
years ago is speculative, Green said, but
"it's better than what we could have
said before."
|