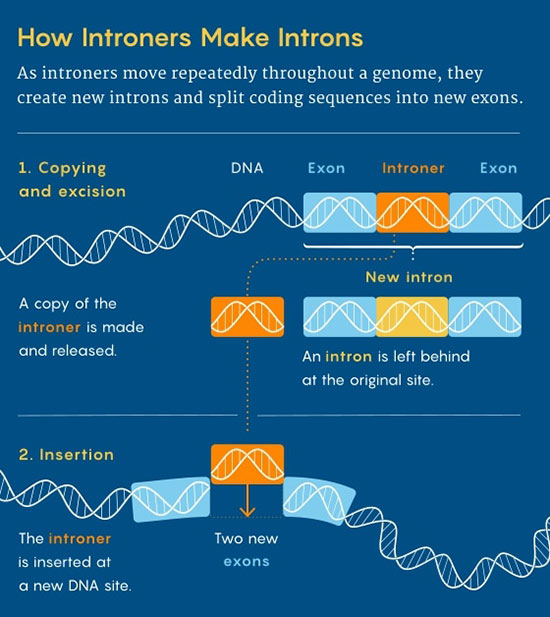
|
by Jake Buehler from QuantaMagazine Website
into
the middle of brightly colored DNA in a double helix. in the genes of complex organisms may have been inserted there
by
parasitic mobile genetic elements called introners.
A novel type of "jumping gene" may explain why the genomes of complex cells aren't all equally stuffed with noncoding sequences...
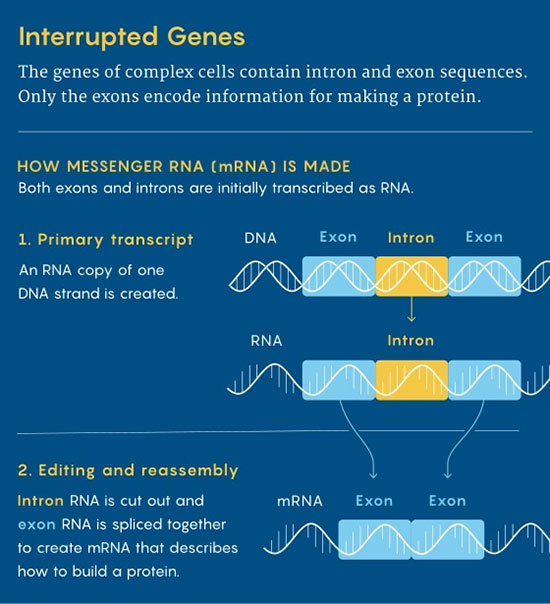
In their DNA, the information about how to make proteins isn't laid out in long coherent strings of bases.
Instead, genes are split into segments, with intervening sequences, or "introns," spacing out the exons that encode bits of the protein.
When eukaryotes express
their genes, their cells have to splice out RNA from the introns and
stitch together RNA from the exons to reconstruct the recipes for
their proteins.
The genes of yeast, for instance, have very few introns, but those of land plants have many.
How this tremendous,
enigmatic variation in intron frequency evolved has stirred debate
among scientists for decades.
A 3D model of a
spliceosome. perform the vital job in complex cells of removing the intron RNA from genes being transcribed and assembling
the other segments into messenger RNA.
These pieces of DNA can slip into genomes and multiply there, leaving profusions of introns behind them. Last November, researchers presented evidence that introners have been doing this in diverse eukaryotes throughout evolution.
Moreover, they showed
that introners could explain why explosive gains in introns seem to
have been particularly common in aquatic forms of life.
For that reason, all eukaryotic cells are equipped with special genetic shears called spliceosomes.
These protein complexes recognize the distinctive sequences that flank intron RNA and remove it from the preliminary RNA transcripts of active genes.
Then they splice together
the coding segments from exons to produce messenger RNA that can be
translated into a working protein.
Quanta Magazine
But the key might be that such introns allow for alternative splicing, a phenomenon that dramatically increases the diversity of products that can arise from a single gene.
When the intron RNA is
clipped out, the exon RNA sequences can be strung together in a new
order to make slightly different proteins, Corbett-Detig explained.
Since the discovery of introns in 1977, researchers have developed numerous theories about where these intrusive sequences came from.
Several mechanisms that
could create introns have been identified, and all of them may have
contributed some introns to eukaryotes. But it's been hard to say
which if any of them might explain where the majority of introns
came from.
Some lineages are particularly heavy with them in ways that point to sudden inundations with introns during their evolutionary history.
When you examine the tree of life and how many introns are found on each tip of the tree, Corbett-Detig said,
One possible explanation for those explosive infusions of introns involves an unusual kind of genetic element known as an introner.
First described in 2009
in the unicellular green algae Micromonas, introners have
subsequently turned up in the genomes of some other algae, some
species of fungi, tiny marine organisms called dinoflagellates and
simple invertebrates called tunicates.
Then they move on, leaving behind a specific intron sequence flanked by splicing sites, which splits the coding DNA into two exons. This process can be repeated on a massive scale throughout a genome.
In fungi, for example,
introners appear to account for most of the intron gain during at
least the last 100,000 years.
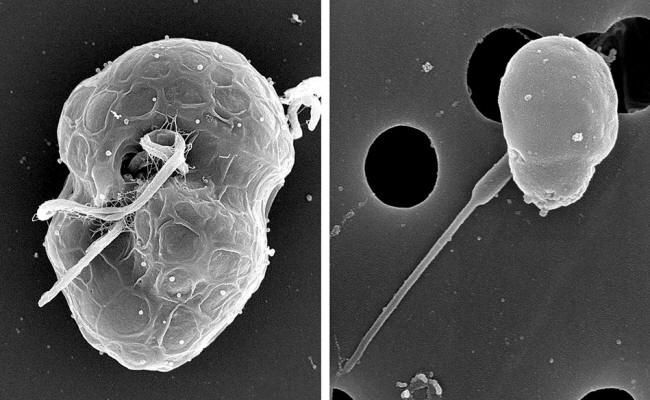
Micrographs of Polarella
and Micromonas. were only known to be in a few organisms, such as the dinoflagellate Polarella glacialis (left)
and the green algae Micromonas. courtesy of Elodie Foulon/Roscoff Culture Collection,
Sorbonne Université and CNRS
Transposons also insert
huge numbers of copies of themselves into genomes.
Introners could cause introns to burst forth in genomes in great numbers, which might explain the punctuated pattern of their emergence in various eukaryotes.
The catch was that introners were only known to exist in a few organisms.
A look at the scientific literature showed that no groups had published any data about introners elsewhere among the eukaryotes.
Gozashti, now at Harvard University, Corbett-Detig and their colleagues set out to remedy that.
They used a series of computational filters to identify potential introners, looking for introns with very similar sequences and whittling away false positives.
In the end, they found
thousands of introns derived from introners in 175 of those genomes,
about 5% of the total, from 48 different species.
The evolutionary lineages of many species alive today may have experienced floods of introns, but any influx that occurred more than a few million years ago would be undetectable.
The 5% result therefore
hints that introners may be far more ubiquitous.
Quanta Magazine
A good parasite can't draw too much attention to itself. If an introner disrupts the activity of the gene in which it has embedded itself, it could harm the host organism, and natural selection could remove the genomic parasite altogether.
So these elements are
continually evolving to be "as neutral as possible" in their
influence, said
Valentina Peona, a comparative genomicist at Uppsala
University.
For example, introners
are more than six times as likely to appear in the genomes of
aquatic organisms as in those of terrestrial organisms. Moreover,
nearly three-quarters of the genomes from aquatic species that
contain introners host multiple introner families.
These unorthodox gene
transfers tend to happen in aquatic environments or in instances of
close interspecies association, such as between hosts and parasites,
explained
Saima Shahid, a plant biologist at Oklahoma State
University.
Single-celled organisms paddle around in this stew, so it's easy for them to take up foreign DNA that might be incorporated into their own.
But even much more
complex multicellular species lay their eggs or fertilize them in
the water, creating opportunities for DNA to be transferred into
their lineages.
Photo of the cloud of
sperm surrounding fish that are mating.
could be
exposed to horizontal transfers of mobile genetic elements in the
water.
from the book
'700 sharks into the dark,' Andromède
Editions 2017
In 2020, their work uncovered nearly 1,000 distinct horizontal transfers involving transposons that had occurred in over 300 vertebrate genomes.
The
vast majority of these transfers happened in teleost fish, Gilbert
said.
Terrestrial organisms aren't likely to have the same bursts of introns, Corbett-Detig said, since horizontal transfer occurs far less often among them.
The transferred introns
could persist in genomes for many millions of years as permanent
souvenirs from an ancestral life in the sea and a fateful brush with
a deft genomic parasite.
Defense mechanisms that a genome might use to suppress its inherited burden of transposons might not work on an unfamiliar genetic element arriving by horizontal transfer.
Even if the introners are initially harmful, the researchers
hypothesize that selective pressures could soon tame them by cutting
them out of RNA.
But the discovery of introners' widespread influence does challenge some theories about how genomes - particularly eukaryotic genomes - have evolved.
at the University of California, Santa Cruz, thinks that introners could have created most of the introns that
various eukaryote lineages
have gained.
One example involves a theory of intron evolution developed by Michael Lynch of Arizona State University in 2002. Models suggest that in species with small breeding populations, natural selection can be less efficient at removing unhelpful genes.
Lynch proposed that those
species will therefore tend to build up heaps of nonfunctional
genetic junk in their genomes. In contrast, species with very large
breeding populations should not be gaining many introns at all.
Some marine
protists with
gargantuan breeding populations had hundreds or thousands of
introners. In contrast, introners were rare in animals and absent in
land plants - both groups with much smaller breeding populations.
The parasitic elements are in "constant conflict" with genetic elements that belong to the host, Gozashti explained, because they compete for genomic space.
That raises the question
of what the intron gains meant for the functional biology of the
organisms in which they occurred.
The comparison might help to reveal how influxes of introns could promote the appearance of new genes.
Similarly, Feschotte
thinks that profusions of introns might help drive the evolution of
families of genes that can change rapidly. Stuffed with new introns,
those genes could co-opt the new variability enabled by alternative
splicing.
Gozashti sitting at his office desk
in front of monitors showing his genomic work. now at Harvard University, first heard that introners had been seen in only a few species, he was inspired to search for the genetic elements
more comprehensively
in other organisms.
Editor’s note: Gozashti is a graduate student in the laboratory of Hopi Hoekstra, who serves on the advisory board for Quanta.
Venomous species, for instance, often need to remix the complex cocktails of peptides in their venoms at the genetic level to adapt to different prey or predators.
The ability of the immune system to generate endlessly
diverse molecular receptors also depends on genes that can rearrange
and recombine quickly.
They should be considered,
Finding this piece of the
puzzle would help flesh out the full story of where most of
eukaryotes' introns have come from.
For Gozashti, the discovery of introners in such a wide range of eukaryotes holds a lesson about how to approach fundamental questions about the nature of eukaryotic life:
Studies often focus on the sliver of biodiversity represented by animals and land plants.
But to understand the important patterns of genomic information underlying all life,
|