|

by Krista Conger
August
08, 2019
from
StanfordMedicine Website
|
Krista Conger is a science writer for the medical
school's Office of Communication & Public Affairs.
Email her at
kristac@stanford.edu. |
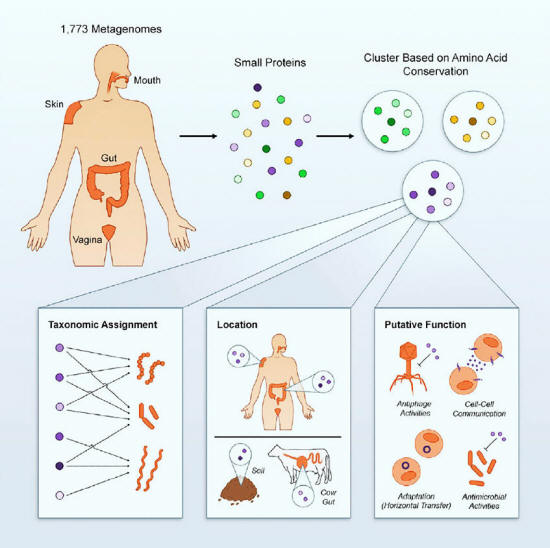
Graphical Abstract
Source
The
bacteria in our gut
make thousands
of tiny, previously unidentified proteins
that could shed
light on human health
and advance drug
development,
Stanford
researchers have found...
Human
Microbiome churns out Thousands of Tiny Novel Proteins
Your body is a wonderland. A wonderland teeming with trillions of
bacteria, that is.
But it's not as
horrifying as it might sound.
In fact, there's mounting
evidence that many aspects of our health are closely intertwined
with the composition and hardiness of our microscopic compatriots,
though exactly how is still mostly unclear.
Now, researchers at the
Stanford University School of Medicine
have discovered that these microbial hitchhikers - collectively
known as the
human microbiome - are churning out
tens of thousands of proteins so small that they've gone unnoticed
in previous studies.
The proteins belong to
more than 4,000 new biological families predicted to be involved in,
among other processes, the warfare waged among different bacterial
strains as they vie for primacy in coveted biological niches, the
cell-to-cell communication between microbes and their unwitting
hosts, and the critical day-to-day housekeeping duties that keep the
bacteria happy and healthy.
Because they are so small - fewer than 50 amino acids in length -
it's likely the proteins fold into unique shapes that represent
previously unidentified biological building blocks.
If the shapes and
functions of these proteins can be recreated in the lab, they could
help researchers advance scientific understanding of how the
microbiome affects human health and pave the way for new drug
discovery.
A paper (Large-Scale
Analyses of Human Microbiomes Reveal Thousands of Small, Novel Genes)
describing the research findings was published Aug. 8 in Cell.
Ami Bhatt, MD, PhD, assistant
professor of medicine and of genetics, is the senior author.
Postdoctoral scholar Hila Sberro, PhD, is the lead author.

Ami Bhatt and her collaborators
found that microbes in the human gut
are making thousands of proteins
so small that they've previously gone undetected.
Norbert von der Groeben
'A Clear Blind
Spot'
"It's critically
important to understand the interface between human cells and
the microbiome," Bhatt said.
"How do they
communicate? How do strains of bacteria protect themselves from
other strains? These functions are likely to be found in very
small proteins, which may be more likely than larger proteins to
be secreted outside the cell."
But the proteins'
miniscule size had made it difficult to identify and study them
using traditional methods.
"We've been more
likely to make an error than to guess correctly when trying to
predict which bacterial DNA sequences contain these very small
genes," Bhatt said.
"So until now, we've
systematically ignored their existence. It's been a clear blind
spot."
The fact that
she found
thousands of new protein families
definitely
surprised us all...
It might be intimidating for the uninitiated to think too deeply
about the vast numbers of bacteria that live on and in each of us.
They account for far more cells in and on the human body than actual
human cells do.
Yet these tiny passengers are rarely malicious.
Instead, they help with
our digestion, supplement our diet and generally keep us running at
our peak. But in many cases, it's been difficult to pick apart the
molecular minutiae behind this partnership.
Bhatt and her colleagues wondered if answers might be found in the
small proteins they knew were likely to wiggle through the nets cast
by other studies focusing on the microbiome.
Small proteins, they
reasoned, are more likely than their larger cousins to slip through
the cell membrane to ferry messages - or threats - to neighboring
host or bacterial cells.
But how to identify and
study these tiny Houdinis...?
"The bacterial genome
is like a book with long strings of letters, only some of which
encode the information necessary to make proteins," Bhatt said.
"Traditionally, we
identify the presence of protein-coding genes within this book
by searching for combinations of letters that indicate the
'start' and 'stop' signals that sandwich genes.
This works well
for larger proteins.
But the smaller the
protein, the more likely that this technique yields large
numbers of false positives that muddy the results."
A Big Surprise
To tackle the problem, Sberro decided to compare potential
small-protein-coding genes among many different microbes and
samples.
Those that were
identified repeatedly in several species and samples were more
likely to be true positives, she thought. When she applied the
analysis to large data sets, Sberro found not the hundreds of genes
she and Bhatt had expected, but tens of thousands.
The proteins predicted to
be encoded by the genes could be sorted into more than 4,000 related
groups, or families, likely to be involved in key biological
processes such as intercellular communication and warfare, as well
as maintenance tasks necessary to keep the bacteria healthy.
"Honestly, we didn't
know what to expect," Bhatt said.
"We didn't have any
intuition about this. The fact that she found thousands of new
protein families definitely surprised us all."
The researchers confirmed
the genes encoded true proteins by showing they are transcribed into
RNA and shuttled to the ribosome for translation - key steps in the
protein-making pathway in all organisms.
They are now working with
collaborators to learn more about the proteins' functions and to
identify those that might be important to the bacteria fighting for
space in our teeming intestinal carpet.
Such proteins might serve
as new antibiotics or drugs for human use, they believe.
"Small proteins can
be synthesized rapidly and could be used by the bacteria as
biological switches to toggle between functional states or to
trigger specific reactions in other cells," Bhatt said.
"They are also easier
to study and manipulate than larger proteins, which could
facilitate drug development. We anticipate this to be a valuable
new area of biology for study."
Bhatt is a member of,
-
Stanford Bio-X
-
the Stanford
Cancer Institute
-
the Stanford
Maternal & Child Health Research Institute
-
a fellow of
Stanford ChEM-H
Other Stanford co-authors
are,
-
graduate student
Brayon Fremin
-
postdoctoral
scholars Soumaya Zlitni, PhD
-
Fredrik Edfors,
PhD
-
Michael Snyder, PhD,
professor and chair of genetics
Researchers from,
-
One Codex
-
the Joint Genome
Institute of the Department of
-
the Alexander
Fleming Biomedical Sciences Research Center in Greece
-
Lawrence Berkeley
National Laboratory,
...also contributed to
the study.
The study was supported by,
-
the National
Institutes of Health (grants HG000044, K08CA184420,
P30CA124435, and 1Ro1AT010123201)
-
the PhRMA
Foundation
-
the U.S.
Department of Energy
-
a Damon Runyon
Clinical Investigator Award
Stanford's departments of
Medicine and of Genetics also supported the work.
|